
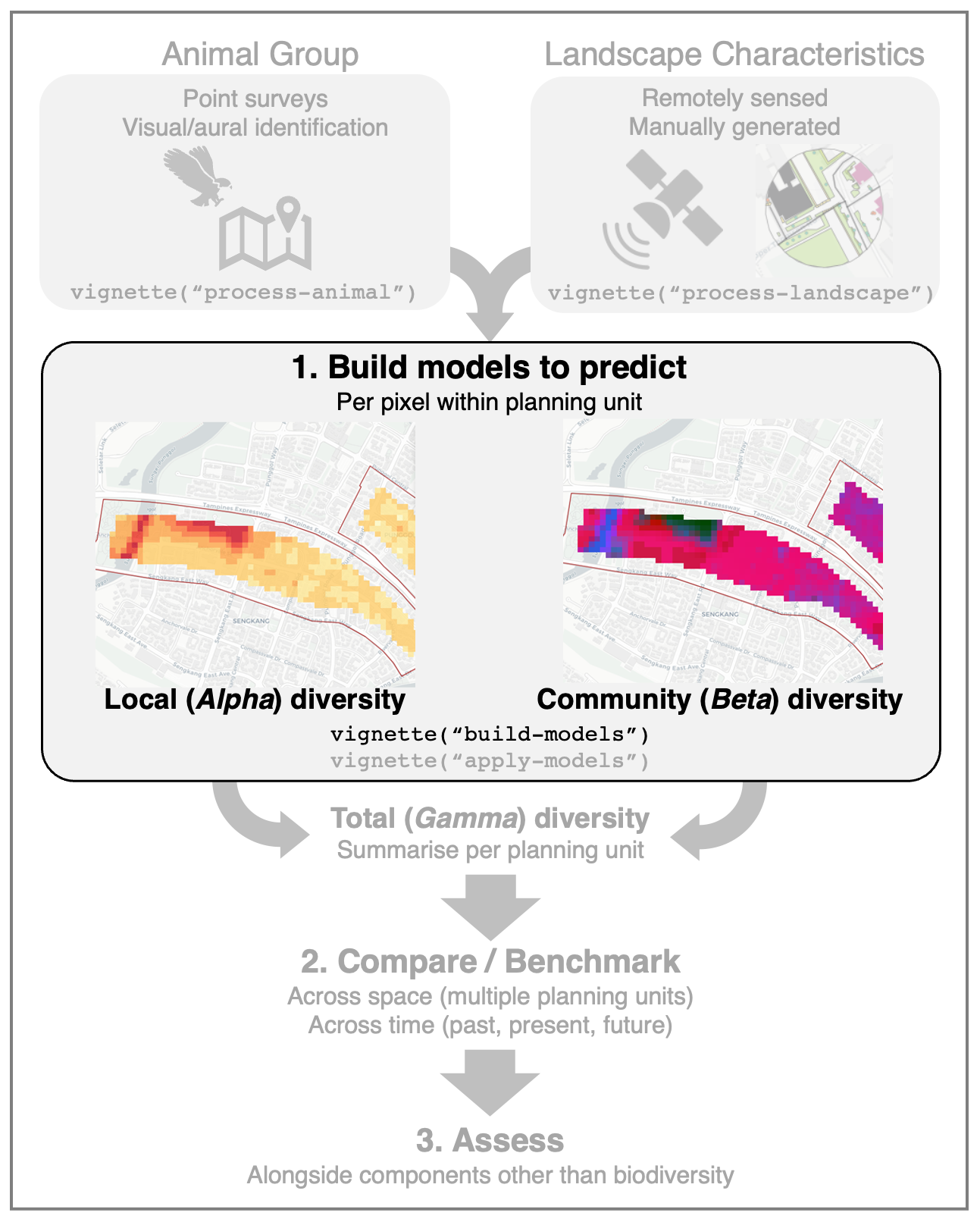
Build Models
Train models that predict animal diversity from landscape data
2022-12-08
Source:vignettes/build-models.Rmd
build-models.RmdThis article outlines a workflow to build predictive models of
diversity for a chosen animal group. Fauna and landscape data should
have already been prepared based on
vignette("process-animal") and
vignette("process-landscape"), respectively. Based on
community ecology theory, we ‘decompose’ animal diversity into two
components: (1) local (Alpha) diversity and (2) community
(Beta) diversity. A predictive model will be built for each
component.
First, load the necessary packages used throughout this article:
library("biodivercity")
library("tidyverse") # to process/wrangle data
library("sf") # to process landscape data1. Local (Alpha) diversity
For a chosen animal group, local (Alpha) diversity is represented by the number of species recorded at sampling point locations (i.e., species density). The response variable is the number (count) of species. Generalised linear mixed-effects models (GLMMs) can be used to model count data, and linear models are generally simpler and provide better interpretability. Do note that other types of models/algorithms can also be used to model count data. Also, the full list of potential predictor variables may first be narrowed down; for example, random forest models may be used to select variables based on their relative importance in improving model performance (Arthur et al., 2010; Li et al., 2017).
Load the necessary packages to model local (Alpha) diversity:
Load the example datasets for birds (animal group ‘Aves’) recorded at
sampling points across Singapore. birds is a dataframe
containing the number of species aggregated across multiple surveys
(i.e., species richness; column ‘sprich’), and
landscape is a dataframe containing landscape predictors
summarised within specified buffer radii. These example datasets can be
obtained by following the steps in vignette(process-animal)
and vignette(process-landscape), respectively. See
data(animal_surveys) and
data(points_landscape) for further details about the
underlying animal and landscape data.
filepath <- system.file("extdata", "build-models_alpha-diversity.Rdata", package="biodivercity")
load(filepath)Pivot the landscape_data to wide format, with the prefix
r<radius>m_ (representing the radius size) appended
to the column names of predictors:
landscape <- landscape %>%
pivot_longer(cols = c(starts_with("osm_"), starts_with("lsm_")),
names_to = 'metric') %>%
pivot_wider(id_cols = c("town", "point_id"),
names_from = c("radius_m", "metric"),
values_from = "value",
names_glue = "r{radius_m}m_{metric}")
# combine bird and landscape data, then then scale/center variables
birds <- birds %>%
inner_join(landscape, by = c("town", "point_id"))
birds_scaled <- birds %>%
mutate(across(.cols = where(is.numeric) & !sprich, scale))
head(birds_scaled) # view combined data## # A tibble: 6 × 20
## town priority point…¹ sprich r200m_…² r200m…³ r200m…⁴ r200m…⁵ r200m…⁶ r200m…⁷
## <chr> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 PG Aves PGT15 14 0.00303 0.575 0.694 -0.497 -0.559 0.456
## 2 PG Aves PGT14 21 0.852 0.344 1.72 -0.485 -0.496 0.621
## 3 PG Aves PGT6 21 1.66 0.997 2.71 -0.511 -0.544 1.01
## 4 QT Aves QTNa14… 34 -1.17 -0.0279 -0.740 3.54 3.40 -0.859
## 5 QT Aves QTNb1a… 40 -1.35 -0.948 -0.799 1.60 1.31 -1.97
## 6 PG Aves PGT7 18 1.33 0.801 2.52 -0.472 -0.490 0.440
## # … with 10 more variables: r200m_lsm_veg_lpi <dbl[,1]>,
## # r200m_lsm_veg_pland <dbl[,1]>, r400m_osm_buildingVol_m3 <dbl[,1]>,
## # r400m_osm_laneDensity <dbl[,1]>, r400m_osm_buildingFA_ratio <dbl[,1]>,
## # r400m_lsm_veg_area_mn <dbl[,1]>, r400m_lsm_veg_gyrate_mn <dbl[,1]>,
## # r400m_lsm_veg_ed <dbl[,1]>, r400m_lsm_veg_lpi <dbl[,1]>,
## # r400m_lsm_veg_pland <dbl[,1]>, and abbreviated variable names ¹point_id,
## # ²r200m_osm_buildingVol_m3[,1], ³r200m_osm_laneDensity[,1], …To fit the model as a GLMM, the function MuMIn::dredge()
will be used. The function includes customisation options; for instance,
the subset argument allows the user to exclude certain
combinations of variables within the fitted model (i.e., apply
constraints to possible models that may be built). In the following
example, we do not allow the same landscape predictor summarised within
multiple buffer radii to be present in the same model.
predictors <- grep("^r[0-9]+m_", colnames(birds_scaled), value = TRUE) # get predictor names
# get rows with same predictors but different radii
var_comb <- combn(predictors, 2) %>% # unique combinations
t() %>%
as.data.frame() %>% # separate radii & predictors
separate(V1, c("V1_radius", "V1_predictor"),
sep = "(?<=[0-9])m_(?=[A-Za-z]*)", remove = FALSE) %>%
separate(V2, c("V2_radius", "V2_predictor"),
sep = "(?<=[0-9])m_(?=[A-Za-z]*)", remove = FALSE) %>%
rowwise() %>%
mutate(similar = grepl(V1_predictor, V2_predictor)) %>%
filter(isTRUE(similar))
# create object to be supplied to function argument
subset_vect <- paste(var_comb$V1, "&", var_comb$V2)
subset_exp <- parse(text = paste0("!(", paste(subset_vect, collapse = ") & !("), ")")) # function argument requires an expressionNext, we fit both a global (maximal) model and null model, in order
to calculate the difference in Akaike Information Criterion (AIC) value
within the function MuMIn::dredge(). In the example data,
the ‘town’ is specified as the random effect. The argument
‘family’ is specified as "poisson", since the
response variable is count data. See the function
lme4::glmer() for more details and information on its
arguments.
model_global <-
glmer(paste("sprich", "~", paste(predictors, collapse = " + "),
"+ (1|town) "),
family = "poisson",
na.action = "na.fail",
control = lme4::glmerControl(optimizer = "bobyqa"),
data = birds_scaled)
model_null <-
glmer(sprich ~ 1 + (1|town), family = "poisson", data = birds_scaled)Automated model selection is performed using the function
MuMIn::dredge(). Models are ranked based on their
AICc value (AIC adjusted for small sample sizes). Model
constraints are specified in the argument subset. To avoid
a long runtime in this demonstration, we also limit the number of
predictor variables in each model to four (argument
m.lim).
model_glm <- dredge(model_global,
subset= subset_exp,
m.lim=c(NA,4), # max of 4 variables
rank="AICc")The resulting object contains a model selection table, with each row representing a model that was fit. It is arranged in ascending order, based on AIC value (lower AIC value represents a ‘better’ model). A summary of the top models with values of ΔAIC < 2 can be viewed:
bestmodels_info <- subset(model_glm,
delta < 2,
recalc.weights=FALSE)
knitr::kable(bestmodels_info, caption = "**Summary of best models ranked based on automated model selection from `MuMIn::dredge()` (ΔAIC < 2).**") %>%
kableExtra::kable_styling("striped") %>% kableExtra::scroll_box(width = "100%", height = "300px")| (Intercept) | r200m_lsm_veg_area_mn | r200m_lsm_veg_ed | r200m_lsm_veg_gyrate_mn | r200m_lsm_veg_lpi | r200m_lsm_veg_pland | r200m_osm_buildingFA_ratio | r200m_osm_buildingVol_m3 | r200m_osm_laneDensity | r400m_lsm_veg_area_mn | r400m_lsm_veg_ed | r400m_lsm_veg_gyrate_mn | r400m_lsm_veg_lpi | r400m_lsm_veg_pland | r400m_osm_buildingFA_ratio | r400m_osm_buildingVol_m3 | r400m_osm_laneDensity | df | logLik | AICc | delta | weight | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 197 | 3.196216 | NA | NA | 0.0551637 | NA | NA | NA | -0.1359401 | -0.0850794 | NA | NA | NA | NA | NA | NA | NA | NA | 5 | -365.8589 | 742.2395 | 0.0000000 | 0.05939683 |
| 194 | 3.196120 | 0.0514166 | NA | NA | NA | NA | NA | -0.1412017 | -0.0832377 | NA | NA | NA | NA | NA | NA | NA | NA | 5 | -366.1590 | 742.8397 | 0.6002541 | 0.04399666 |
| 199 | 3.195786 | NA | -0.0256461 | 0.0461363 | NA | NA | NA | -0.1280837 | -0.0802518 | NA | NA | NA | NA | NA | NA | NA | NA | 6 | -365.3751 | 743.4871 | 1.2475980 | 0.03183104 |
| 709 | 3.196645 | NA | NA | 0.0488160 | NA | NA | NA | -0.1267322 | -0.0812710 | NA | -0.0231842 | NA | NA | NA | NA | NA | NA | 6 | -365.4697 | 743.6763 | 1.4368451 | 0.02895718 |
| 8389 | 3.195210 | NA | NA | 0.0542352 | NA | NA | NA | -0.1271918 | -0.0833486 | NA | NA | NA | NA | NA | -0.0209911 | NA | NA | 6 | -365.5583 | 743.8534 | 1.6139420 | 0.02650332 |
| 205 | 3.195503 | NA | NA | 0.0447909 | 0.0269169 | NA | NA | -0.1262190 | -0.0771083 | NA | NA | NA | NA | NA | NA | NA | NA | 6 | -365.5612 | 743.8592 | 1.6196856 | 0.02642731 |
| 229 | 3.195477 | NA | NA | 0.0553025 | NA | NA | -0.0183244 | -0.1260268 | -0.0838809 | NA | NA | NA | NA | NA | NA | NA | NA | 6 | -365.6520 | 744.0408 | 1.8013640 | 0.02413249 |
| 196 | 3.195746 | 0.0419566 | -0.0251969 | NA | NA | NA | NA | -0.1331441 | -0.0792678 | NA | NA | NA | NA | NA | NA | NA | NA | 6 | -365.7112 | 744.1592 | 1.9197703 | 0.02274524 |
| 706 | 3.196583 | 0.0449303 | NA | NA | NA | NA | NA | -0.1311233 | -0.0796107 | NA | -0.0241621 | NA | NA | NA | NA | NA | NA | 6 | -365.7387 | 744.2143 | 1.9747749 | 0.02212821 |
We can also visualise the importance (based on sum-of-weights) and effect direction (based on coefficient value) for each variable within the top models:
# average the effect of each predictor
effects <- data.frame(coef(bestmodels_info)[,-1]) %>%
summarise(across(everything(), ~ mean(.x, na.rm = TRUE))) %>%
pivot_longer(everything(), names_to = "var") %>%
mutate(effect = ifelse(value > 0, "Positive", "Negative"))
# get importance of each predictor
importances <- data.frame(MuMIn::sw(bestmodels_info)) %>%
rename(imp = MuMIn..sw.bestmodels_info.) %>%
rownames_to_column("var") %>%
left_join(effects, by = "var") %>% # join with effects
mutate(effect = ifelse(is.na(effect), "Mixed (factor)", effect)) %>%
mutate(effect = factor(effect, levels = c("Positive", "Negative", "Mixed (factor)")))
# plot
ggplot(data=importances, aes(x = imp, y = reorder(var, imp),
fill = effect)) +
geom_bar(stat = "identity") +
labs(x = "Sum of weights", y = "Variables") +
scale_fill_manual(values = c("#4daf4a", "#e41a1c", "#377eb8"),
name = "Effect")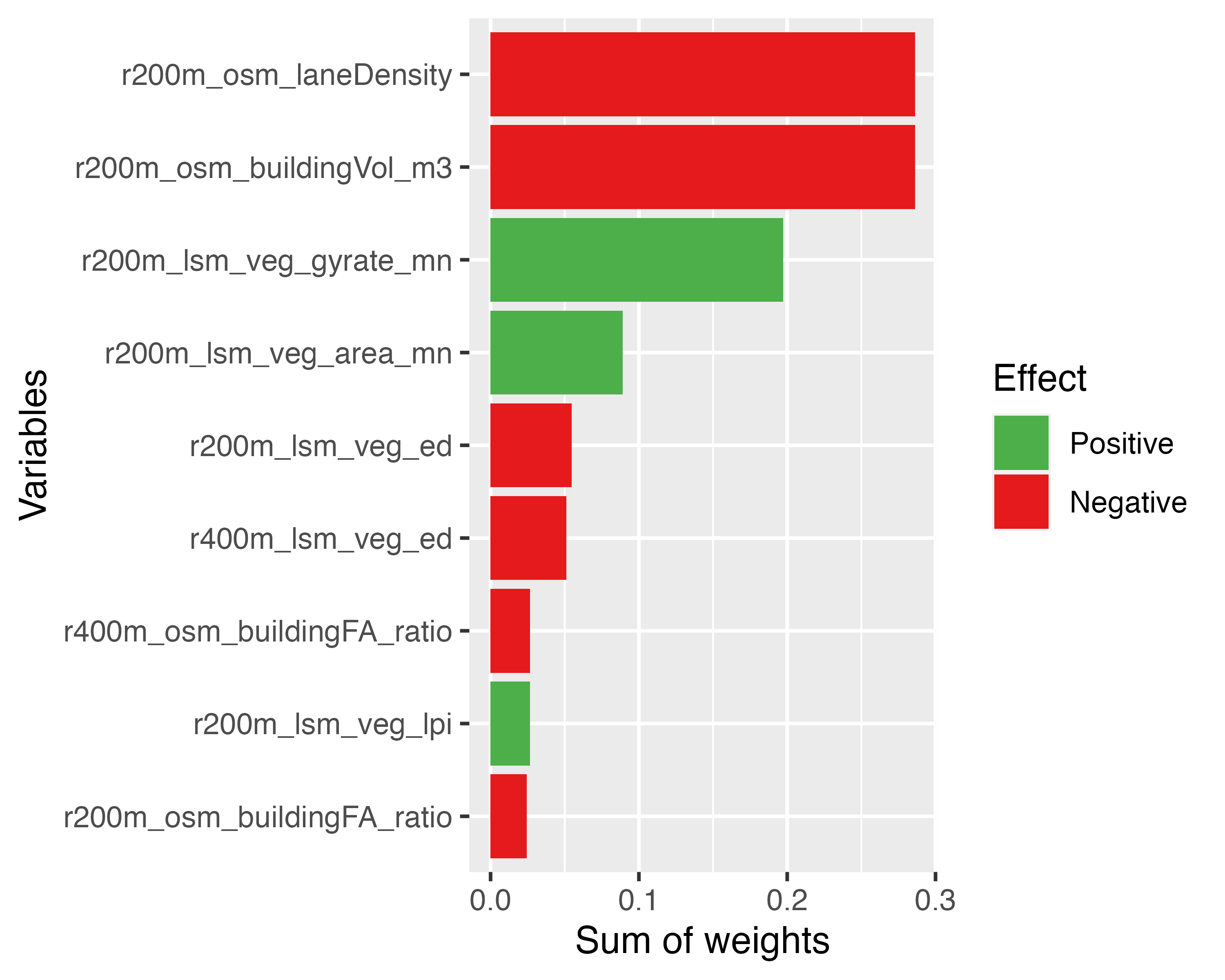
Finally, we can extract the models and recipes::recipe()
objects for future use:
# get list of best models
bestmodels <- MuMIn::get.models(bestmodels_info, subset = TRUE)
# create recipes object only with predictors within best models
predictors_best <- colnames(coef(bestmodels_info)[,-1])
birds <- birds %>% # original unscaled dataset
dplyr::select(sprich, town, all_of(predictors_best)) # remove unnecessary columns
recipe_birds <- recipes::recipe(birds) %>%
recipes::update_role(sprich, new_role = "outcome") %>%
recipes::update_role(town, new_role = "id") %>%
recipes::update_role_requirements("id", bake = FALSE) %>%
recipes::step_normalize(all_of(predictors_best)) %>%
recipes::prep()References
Arthur AD, Li J, Henry S & Cunningham SA (2010) Influence of woody vegetation on pollinator densities in oilseed Brassica fields in an Australian temperate landscape. Basic and Applied Ecology, 11(5): 406–414.
Li J, Alvarez B, Siwabessy J, Tran M, Huang Z, Przeslawski R, Radke L, Howard F & Nichol S (2017) Application of random forest and generalised linear model and their hybrid methods with geostatistical techniques to count data: Predicting sponge species richness. Environmental Modelling and Software, 97: 112–129.