
Animals: Species-level Analyses
Examine survey data at the level of individual species
2022-12-08
Source:vignettes/more/animals-species-analyses.Rmd
animals-species-analyses.RmdSurvey data for each animal (taxon) group can be summarised at multiple levels (e.g. area, period). For a particular animal group of interest, finer-grained summaries can also be generated—at the level of individual species. This article outlines how such summaries may be generated, using bird surveys conducted in Singapore as an example.
First, load the necessary packages to run the analysis:
Prepare data
Load the example data. Refer to
help(animal_observations) and
help(animal_surveys) for more details on these
datasets.
Show group-level (family/genus) observations where all species in the
group are observed at each sampling point, using the function
check_taxongrps(). These entries will later be removed at
the respective points, in order to prevent over-counting the number of
species. More details in vignette("process-animal").
remove <- check_taxongrps(animal_observations, level = "point")
remove## # A tibble: 0 × 4
## # Groups: point_id, period [0]
## # … with 4 variables: point_id <chr>, period <dbl>, name <chr>, n <int>Most abundant species
Summarise the animal surveys based on how many individuals of each
species are present at each sampling point. Tally the number of
individuals per species, within each area,
period and taxon:
plot_fauna <- animal_observations %>%
group_by(species, area, period, taxon) %>%
summarise(count = sum(abundance)) %>%
mutate(area = factor(area, # re-arrange factors
levels = c("PG", "QT", "TP","JW", "BS", "WL")))
plot_fauna## # A tibble: 1,664 × 5
## # Groups: species, area, period [1,664]
## species area period taxon count
## <chr> <fct> <dbl> <chr> <dbl>
## 1 Accipiter gularis TP 2 Aves 1
## 2 Accipiter spp. JW 2 Aves 1
## 3 Accipiter spp. QT 2 Aves 1
## 4 Accipiter spp. TP 2 Aves 2
## 5 Accipiter trivirgatus QT 1 Aves 1
## 6 Accipitriformes PG 2 Aves 1
## 7 Accipitriformes QT 2 Aves 1
## 8 Accipitriformes TP 1 Aves 1
## 9 Acisoma panorpoides BS 1 Odonata 1
## 10 Acisoma panorpoides JW 2 Odonata 11
## # … with 1,654 more rowsThe results can be visualised, for example, for bird species at the six areas (towns) in Singapore, across two survey periods:
# get the (descending) order of species to be plot
order <- plot_fauna %>%
filter(taxon == "Aves") %>% # birds only
group_by(species) %>%
summarise(order = sum(count))
plot_fauna %>%
filter(taxon == "Aves") %>% # birds only
group_by(area, period) %>% # only plot top 10 species
slice_max(count, n = 10) %>%
left_join(order) %>% # get the order
ggplot(aes(y = count, x = reorder(species, count))) +
geom_col(width = 0.8, color = "black") +
facet_grid(~ area + period) +
ylab("Number of individuals") +
xlab("")+
theme_bw()+
theme(text = element_text(size=8.0),
axis.text.y = element_text(face = "italic"),
panel.grid.major.y = element_blank(),
panel.grid.minor.x = element_blank()) +
coord_flip() +
scale_y_continuous(expand = c(0,0), breaks = scales::breaks_pretty(n = 3))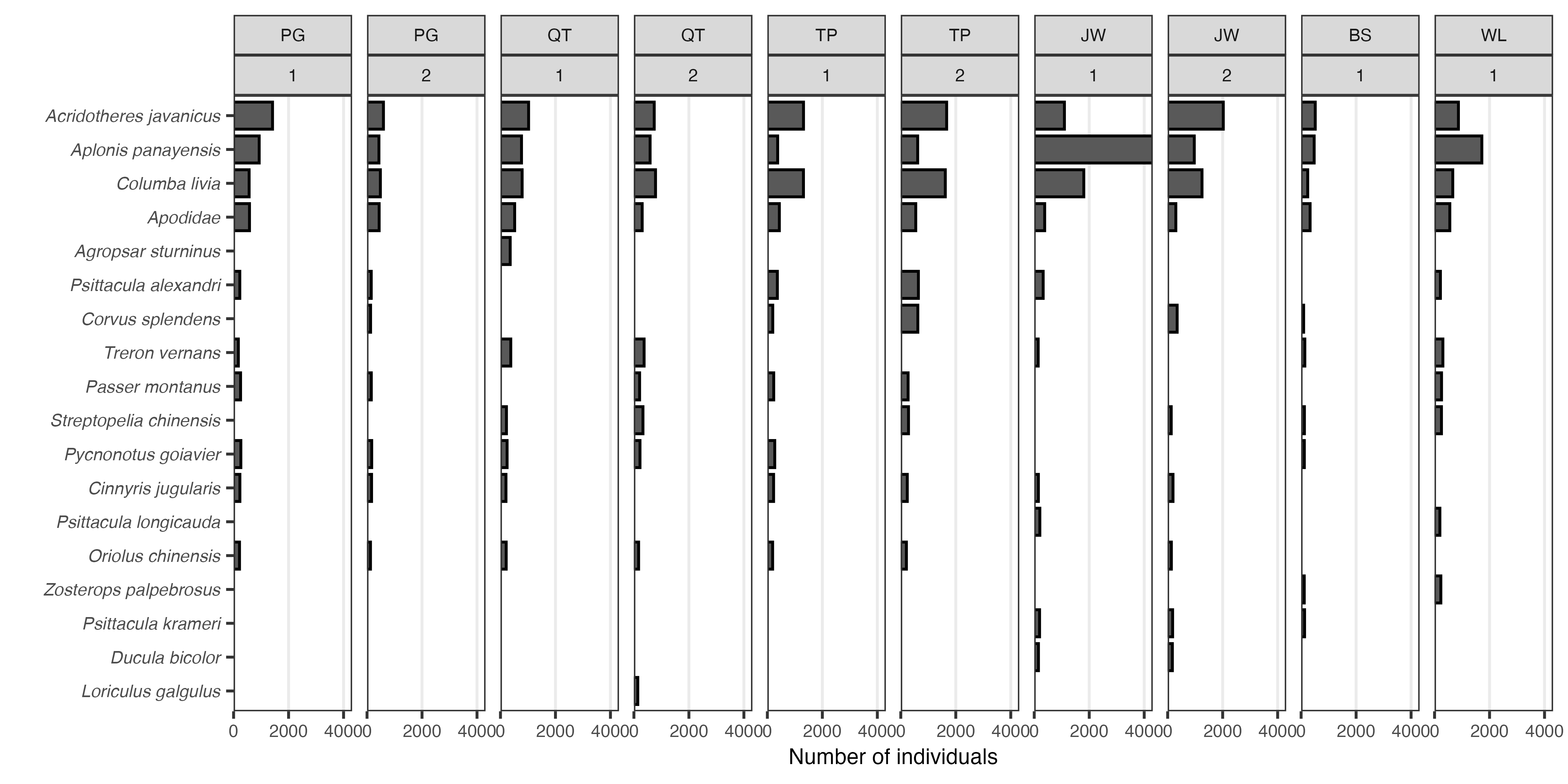
Figure: Most abundant bird species within each area (town), per survey round. Up to ten species are shown; species are arranged in descending order according to their cumulative counts.
Most common species
Summarise the animal surveys based on whether each species is present
at sampling points. Tally the instances where each species
is observed within each area, period,
taxon and point_id:
plot_fauna <- animal_observations %>%
group_by(species, area, period, taxon, point_id) %>% # counts of unique species at each sampling pt, per survey area/period/taxon
summarise() %>%
anti_join(remove, by = c("species" = "name","point_id", "period")) %>% # exclude genus/family lvl records if all species within grp observed
group_by(species, area, period, taxon) %>% # no. of points that each species occurs (per area/period/taxon)
summarise(count = n()) %>%
mutate(area = factor(area, # re-arrange factors
levels = c("PG", "QT", "TP","JW", "BS", "WL"))) Similarly, common bird species within the six areas (towns) in Singapore can be visualised:
# get the (descending) order of species to be plot
order <- plot_fauna %>%
filter(taxon == "Aves") %>% # birds only
group_by(species) %>%
summarise(order = sum(count))
plot_fauna %>%
filter(taxon == "Aves") %>% # birds only
group_by(area, period) %>% # only plot top 10 species
slice_max(count, n = 10) %>%
left_join(order) %>% # get order
ggplot(aes(y = count, x = reorder(species, order))) +
geom_col(width = 0.8, color = "black") +
facet_grid(~ area + period) +
ylab("Number of sampling points") +
xlab("")+
theme_bw()+
theme(text = element_text(size=8.0),
axis.text.y = element_text(face = "italic"),
panel.grid.major.y = element_blank(),
panel.grid.minor.x = element_blank()) +
coord_flip() +
scale_y_continuous(expand = c(0,0), breaks = scales::breaks_pretty(n = 4))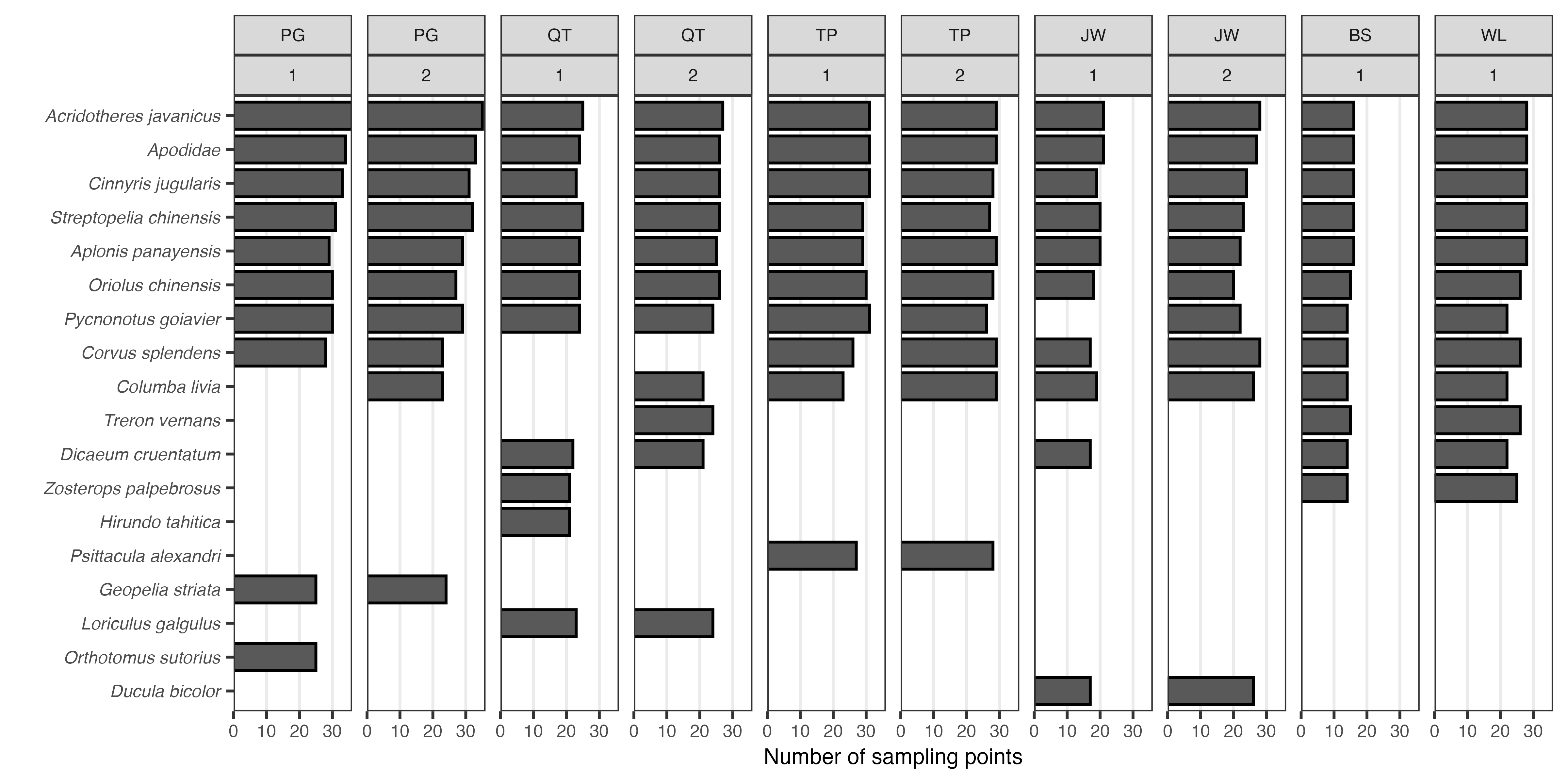
Figure: Most common bird species within each area (town), per survey round. Up to ten species are shown; species are arranged in descending order according to the cumulative number of sampling points they are present in.
More interesting visualisations can also be generated. For example, a hierarchical tree map may be used to show the ten most common bird species in the first survey period:
library(treemapify)
plot_fauna %>%
filter(taxon == "Aves" & period == 1) %>% # birds in period 1 only
group_by(area) %>% # only plot top 10 species
slice_max(count, n = 10) %>%
ggplot(aes(area = count, fill = area, subgroup = species)) +
geom_treemap() +
geom_treemap_subgroup_border(color = "white", size = 7) +
geom_treemap_subgroup_text(place = "centre",
size = 16,
reflow = TRUE, # wrap text
fontface = "bold") +
colorspace::scale_fill_discrete_qualitative("Harmonic") +
labs(title="Urban birds of Singapore",
caption="Most common spp. during surveys in 2016–2019",
fill = NULL) +
theme(aspect.ratio = 5/4,
plot.title = element_text(hjust = 0.5, size = 30, face = "bold"),
plot.caption = element_text(size = 12, face = "italic"),
legend.position="bottom",
legend.text = element_text(size = 12)) +
guides(fill = guide_legend(nrow = 1))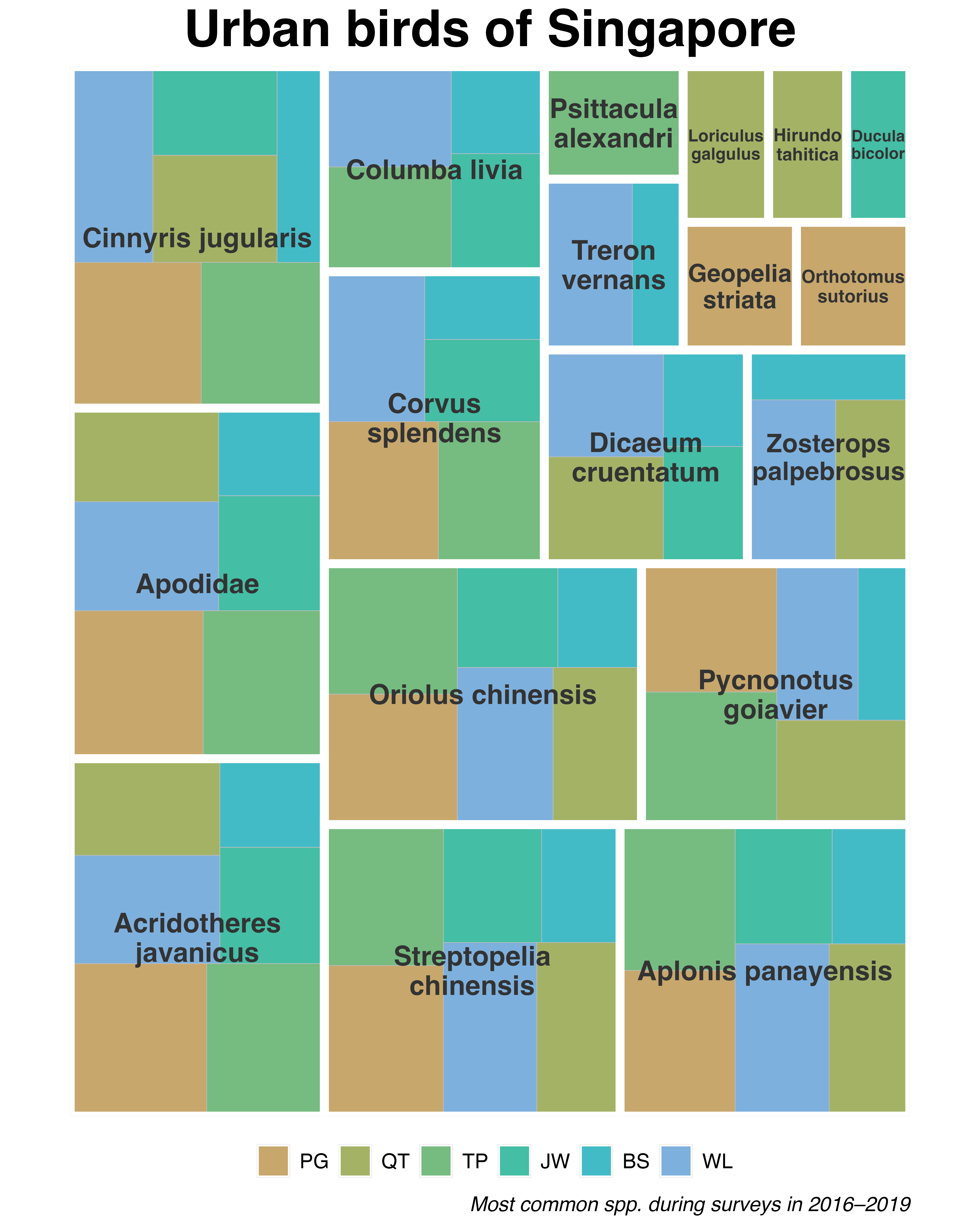
References
The latest accepted taxonomic names and local statuses in the example data are based on the following sources:
- Birds: ebird Clements Checklist; The Singapore Red Data Book (Davison et al. 2008)
- Butterflies: A Field Guide to the Butterflies of Singapore (Khew, 2010)
- Odonates: The Dragonflies of Singapore: an updated checklist and revision of the national conservation statuses (Ngiam & Cheong, 2016)
- Amphibians: A Guide to the Amphibians & Reptiles of Singapore (Lim & Lim, 2002)