
Process Animal Data
Format, check and summarise data from animal surveys conducted at point locations
2022-12-08
Source:vignettes/process-animal.Rmd
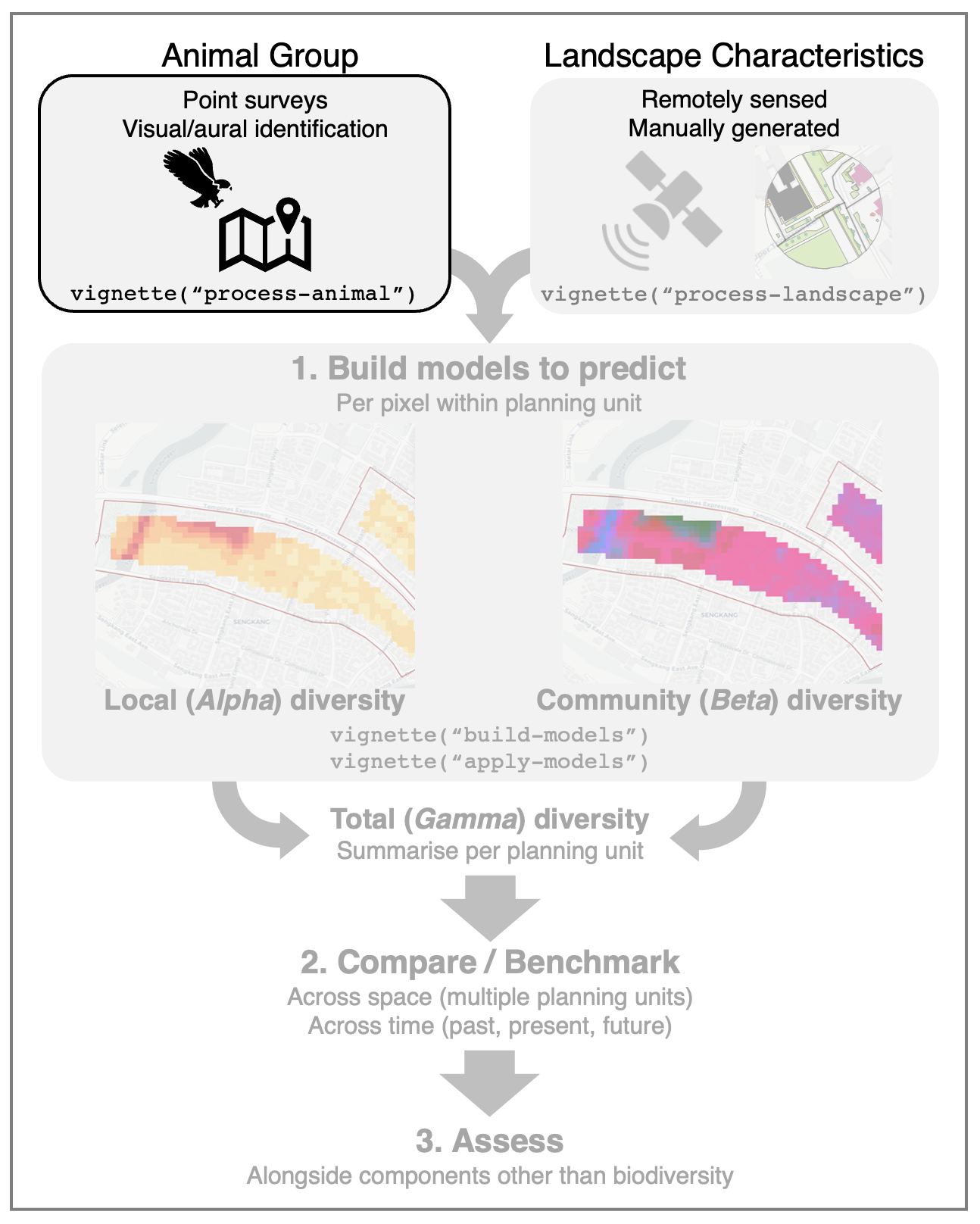
process-animal.RmdAnimal observations from on-site surveys can be used to build predictive models of animal diversity, or summarised to directly compare animal diversity between different areas or time periods. This article outlines the ways to format, check and process the animal data.
First, load the necessary packages to run the analysis:
library("biodivercity")
library("dplyr") # data processingData format
Data from animal surveys are organised into two separate tables: (1)
a record of animal observations during each survey; and (2) reference
information about each survey. The existence of (2) ensures that surveys
with zero animal observations are accounted for. More details on how
data are collected can be found in
vignette("animals-survey-protocols"). These example data
can be loaded by running the following code:
| survey_id | point_id | area | period | cycle | resampled | start_time | time | taxon | species | family | genus | abundance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 QTNa14a_P 1 Odonata | QTNa14a_P | QT | 1 | 1 | NA | 2016-08-04 14:00:00 | 2016-08-04 14:01:00 | Odonata | Rhyothemis phyllis | Libellulidae | Rhyothemis spp. | 3 |
| 1 QTNa14a_P 1 Odonata | QTNa14a_P | QT | 1 | 1 | NA | 2016-08-04 14:00:00 | 2016-08-04 14:12:00 | Odonata | Crocothemis servilia | Libellulidae | Crocothemis spp. | 1 |
| 1 QTNa14a_P 1 Odonata | QTNa14a_P | QT | 1 | 1 | NA | 2016-08-04 14:00:00 | 2016-08-04 14:15:00 | Odonata | Neurothemis fluctuans | Libellulidae | Neurothemis spp. | 1 |
| 1 QTNa14a_P 1 Odonata | QTNa14a_P | QT | 1 | 1 | NA | 2016-08-04 14:00:00 | 2016-08-04 14:23:00 | Odonata | Crocothemis servilia | Libellulidae | Crocothemis spp. | 1 |
| 1 QTNa14a_P 1 Odonata | QTNa14a_P | QT | 1 | 1 | NA | 2016-08-04 14:00:00 | 2016-08-04 14:28:00 | Odonata | Neurothemis fluctuans | Libellulidae | Neurothemis spp. | 1 |
| 1 QTNb1a_P 1 Odonata | QTNb1a_P | QT | 1 | 1 | NA | 2016-08-04 14:44:00 | 2016-08-04 14:44:00 | Odonata | Neurothemis fluctuans | Libellulidae | Neurothemis spp. | 1 |
| survey_id | point_id | area | period | cycle | taxon | resampled | start_time | notes |
|---|---|---|---|---|---|---|---|---|
| 1 QTNa14a_P 1 Odonata | QTNa14a_P | QT | 1 | 1 | Odonata | NA | 2016-08-04 14:00:00 | NA |
| 1 QTNb1a_P 1 Odonata | QTNb1a_P | QT | 1 | 1 | Odonata | NA | 2016-08-04 14:44:00 | NA |
| 1 QTNa14a_P 1 Amphibia | QTNa14a_P | QT | 1 | 1 | Amphibia | NA | 2016-08-04 19:55:00 | NA |
| 1 QTNb1a_P 1 Amphibia | QTNb1a_P | QT | 1 | 1 | Amphibia | NA | 2016-08-04 20:40:00 | NA |
| 1 PGT15 1 Aves | PGT15 | PG | 1 | 1 | Aves | NA | 2016-08-08 07:00:00 | NA |
| 1 PGT14 1 Aves | PGT14 | PG | 1 | 1 | Aves | NA | 2016-08-08 07:33:00 | NA |
If necessary, the convenience function
filter_observations() may be used to filter the animal
survey data based on specified grouping variables
(e.g. area, period, taxon). For
example, observations from bird (Aves) surveys in Tampines
(TP) conducted during 2020-2021 (survey period
2) can be filtered as follows:
observations_subset <- filter_observations(observations = animal_observations,
survey_ref = animal_surveys,
specify_taxon = "Aves",
specify_area = "TP",
specify_period = "2")
observations_subset## # A tibble: 4,180 × 13
## survey_id point_id area period cycle resampled start_time
## <fct> <fct> <chr> <dbl> <dbl> <lgl> <dttm>
## 1 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## 2 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## 3 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## 4 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## 5 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## 6 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## 7 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## 8 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## 9 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## 10 2 TPa3b 1 Aves TPa3b TP 2 1 FALSE 2020-07-01 06:55:00
## # … with 4,170 more rows, and 6 more variables: time <dttm>, taxon <chr>,
## # species <chr>, family <chr>, genus <chr>, abundance <dbl>Note that the column survey_id will be converted to a
factor variable, which includes levels that are not present in the
observation data (i.e., no butterflies were observed during those
surveys).
Data checks
Checks can be performed to remove group-level observations, to prevent over-counting the number of species when summarising the data afterwards. For instance, if the species name of an observed animal is unknown, surveyors may have recorded such observations at a higher (group) level of classification, such as the family or genus. However, this same species recorded as a group-level entry may also be identified correctly (at the species-level) during other surveys at the same point/area (e.g., by another surveyor). When the number of species is tallied, it may result in a double-count of the particular animal species.
For example, there are two species of swallows observed across all
points in the example dataset–Hirundo tahitica and Hirundo
rustica. Hirundo spp. is entered (in the
species column) when the species cannot be identified with
confidence:
observations_swallows <- observations_subset %>%
filter(grepl("Hirundo", species)) %>%
group_by(area, point_id, species, family, genus) %>%
summarise()
observations_swallows## # A tibble: 42 × 5
## # Groups: area, point_id, species, family [42]
## area point_id species family genus
## <chr> <fct> <chr> <chr> <chr>
## 1 TP TPa3b Hirundo rustica Hirundinidae Hirundo spp.
## 2 TP TPa3b Hirundo tahitica Hirundinidae Hirundo spp.
## 3 TP TPa15a Hirundo rustica Hirundinidae Hirundo spp.
## 4 TP TPa15a Hirundo spp. Hirundinidae Hirundo spp.
## 5 TP TPa68a_P Hirundo rustica Hirundinidae Hirundo spp.
## 6 TP TPa68a_P Hirundo tahitica Hirundinidae Hirundo spp.
## 7 TP TPa70_P Hirundo rustica Hirundinidae Hirundo spp.
## 8 TP TPa70_P Hirundo tahitica Hirundinidae Hirundo spp.
## 9 TP TPa16a Hirundo spp. Hirundinidae Hirundo spp.
## 10 TP TPa16a Hirundo tahitica Hirundinidae Hirundo spp.
## # … with 32 more rowsThis would inflate the tallied number of species per point/area,
since Hirundo spp. is a unique entry in the
species column. For example, in the example dataset, a
total of three swallow species would be reported within the Tampines
(TP) area, rather than the two known to be present in the
city of Singapore:
observations_swallows %>%
group_by(area, species) %>% # tally by area
summarise() %>%
summarise(n())## # A tibble: 1 × 2
## area `n()`
## <chr> <int>
## 1 TP 3One way to avoid such double-counting is to remove these group-level
entries if all species in that particular classification group are
observed, at the specified granularity of interest (point
or area). The function check_taxongrps()
identifies such group-level entries, as well as the total number of
species within that grouping level (by referring to the columns
genus and family). For example, for the
example dataset of swallows within the Tampines (TP) area,
we can check the number of unique species within the Hirundo
spp. genus and Hirundinidae family:
to_remove <- check_taxongrps(observations_swallows, level = "area")
to_remove## # A tibble: 0 × 3
## # Groups: area [0]
## # … with 3 variables: area <chr>, name <chr>, n <int>These entries can subsequently be removed from
observations_swallows, if they have been recorded in the
species column:
filtered_observations <- observations_swallows %>%
anti_join(to_remove, by = c("species" = "name"))
filtered_observations## # A tibble: 42 × 5
## # Groups: area, point_id, species, family [42]
## area point_id species family genus
## <chr> <fct> <chr> <chr> <chr>
## 1 TP TPa3b Hirundo rustica Hirundinidae Hirundo spp.
## 2 TP TPa3b Hirundo tahitica Hirundinidae Hirundo spp.
## 3 TP TPa15a Hirundo rustica Hirundinidae Hirundo spp.
## 4 TP TPa15a Hirundo spp. Hirundinidae Hirundo spp.
## 5 TP TPa68a_P Hirundo rustica Hirundinidae Hirundo spp.
## 6 TP TPa68a_P Hirundo tahitica Hirundinidae Hirundo spp.
## 7 TP TPa70_P Hirundo rustica Hirundinidae Hirundo spp.
## 8 TP TPa70_P Hirundo tahitica Hirundinidae Hirundo spp.
## 9 TP TPa16a Hirundo spp. Hirundinidae Hirundo spp.
## 10 TP TPa16a Hirundo tahitica Hirundinidae Hirundo spp.
## # … with 32 more rowsTo verify that these entries have been removed, we can re-tally the filtered observations. The number of species will be reduced by one:
filtered_observations %>%
group_by(area, species) %>% # tally by area
summarise() %>%
summarise(n())## # A tibble: 1 × 2
## area `n()`
## <chr> <int>
## 1 TP 3Summarise data
To build predictive models for local (alpha) diversity for a chosen
animal group, the animal observations will need to be aggregated at the
level of each sampling point. For example, we can tally the number of
bird (animal group ‘Aves’) species per point using the function
tally_observations(). Note that this function avoids
double-counting group-level records, by acting as a wrapper to the
function check_taxongrps() (see previous section).
birds <-
tally_observations(observations = animal_observations,
survey_ref = animal_surveys,
level = "point",
specify_taxon = "Aves")
head(birds)## # A tibble: 6 × 5
## area period taxon point_id n
## <chr> <dbl> <chr> <chr> <int>
## 1 BS 1 Aves BSa11a_P 38
## 2 BS 1 Aves BSa13_P 36
## 3 BS 1 Aves BSa14a 35
## 4 BS 1 Aves BSa1a 18
## 5 BS 1 Aves BSa2 16
## 6 BS 1 Aves BSa20a_E 31To build predictive models of community (Beta) diversity for
a chosen animal group, a presence/absence community matrix will need to
be created. For each species (columns) at each sampling point (rows),
presence is denoted as 1 while absence is denoted as
0. For example, we can generate the community matrix for
the animal group ‘Aves’ (birds), after manually removing genus/family
records using the function check_taxongrps():
# manually exclude genus/family lvl records by point
to_remove <-
check_taxongrps(animal_observations, level = "point")
filtered_observations <- animal_observations %>%
anti_join(to_remove, by = c("species" = "name",
"point_id",
"period"))
# generate community matrix
bird_com <- filtered_observations %>%
filter(taxon == "Aves") %>%
group_by(point_id, species) %>% # tally no. of individuals per point and species
summarise(n = sum(abundance)) %>%
group_by(point_id) %>%
pivot_wider(names_from = species, # pivot to wide format
values_from = n) %>%
replace(is.na(.),0) %>%
ungroup() %>%
as.data.frame() %>%
dplyr::select(-point_id) %>%
mutate(across(.cols = everything(), # change to presence/absence
~ case_when(. > 0 ~ 1,
. == 0 ~ 0))) %>%
select(which(colMeans(.) > 0)) # include species observed at least once
head(bird_com)## Acridotheres javanicus Aegithina tiphia Amaurornis phoenicurus
## 1 1 1 1
## 2 1 1 1
## 3 1 1 1
## 4 1 0 0
## 5 1 0 0
## 6 1 1 0
## Aplonis panayensis Apodidae Ardea purpurea Ardeola spp. Bubulcus ibis
## 1 1 1 1 1 1
## 2 1 1 1 0 0
## 3 1 1 1 0 0
## 4 1 1 0 0 1
## 5 1 1 0 0 0
## 6 1 1 1 0 0
## Butorides striata Cinnyris jugularis Columba livia Copsychus saularis
## 1 1 1 1 1
## 2 1 1 1 1
## 3 0 1 1 1
## 4 0 1 1 0
## 5 0 1 1 0
## 6 0 1 0 1
## Corvus splendens Dicaeum cruentatum Eudynamys scolopaceus Geopelia striata
## 1 1 1 1 1
## 2 1 1 1 1
## 3 1 1 1 1
## 4 1 1 1 0
## 5 1 1 0 0
## 6 0 1 1 1
## Halcyon smyrnensis Haliastur indus Hemiprocne longipennis Hirundo rustica
## 1 1 1 1 1
## 2 0 0 0 0
## 3 1 0 0 1
## 4 0 0 0 0
## 5 0 0 0 0
## 6 1 0 1 1
## Ixobrychus sinensis Lanius cristatus Lonchura punctulata Loriculus galgulus
## 1 1 1 1 1
## 2 1 0 1 1
## 3 0 0 0 1
## 4 0 0 0 1
## 5 0 0 0 0
## 6 0 0 0 1
## Muscicapa dauurica Oriolus chinensis Orthotomus sutorius Psittacula alexandri
## 1 1 1 1 1
## 2 1 1 0 0
## 3 1 1 1 0
## 4 0 1 0 0
## 5 0 1 0 0
## 6 1 1 0 0
## Psittacula krameri Psittacula longicauda Psittaculidae Pycnonotus goiavier
## 1 1 1 1 1
## 2 1 0 0 1
## 3 1 0 0 1
## 4 1 0 0 1
## 5 0 0 1 1
## 6 1 0 0 1
## Rhipidura javanica Streptopelia chinensis Todiramphus chloris Treron vernans
## 1 1 1 1 1
## 2 1 1 1 1
## 3 1 1 1 1
## 4 0 1 0 1
## 5 0 1 0 0
## 6 1 1 1 1
## Yungipicus moluccensis Zosterops palpebrosus Acrocephalus orientalis
## 1 1 1 0
## 2 0 1 1
## 3 0 1 0
## 4 0 1 0
## 5 1 1 0
## 6 1 1 0
## Apus nipalensis Ardea cinerea Dinopium javanense Haliaeetus leucogaster
## 1 0 0 0 0
## 2 1 1 1 1
## 3 0 1 1 0
## 4 0 0 1 0
## 5 0 0 0 0
## 6 0 1 0 0
## Lanius schach Orthotomus ruficeps Passer montanus Pelargopsis capensis
## 1 0 0 0 0
## 2 1 1 1 1
## 3 0 0 1 0
## 4 0 0 1 0
## 5 0 0 1 0
## 6 0 1 0 0
## Trichoglossus haematodus Agropsar sturninus Gallus gallus (domestic type)
## 1 0 0 0
## 2 1 0 0
## 3 1 1 1
## 4 0 0 0
## 5 0 0 0
## 6 1 0 1
## Lanius spp. Merops viridis Micropternus brachyurus Anthreptes malacensis
## 1 0 0 0 0
## 2 0 0 0 0
## 3 1 1 1 0
## 4 0 0 0 0
## 5 0 0 0 1
## 6 0 0 0 1
## Hirundo tahitica Phylloscopus borealis Acridotheres tristis Ardeidae
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 1 0 0 0
## 6 1 1 0 0
## Corvus macrorhynchos Falco peregrinus Lalage nigra Orthotomus atrogularis
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Anthus rufulus Dicrurus paradiseus Egretta garzetta Pernis ptilorhynchus
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Ducula bicolor Gerygone sulphurea Dicrurus annectans Ardea intermedia
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Merops philippinus Anthus cervinus Cisticola juncidis Motacilla flava
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Vanellus indicus Actitis hypoleucos Alcedo atthis Lanius tigrinus
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Motacilla cinerea Elanus caeruleus Hirundo spp. Pycnonotus jocosus
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Ardeola bacchus Ardeola speciosa Gallinago spp. Rallidae
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Chrysococcyx minutillus Nectariniidae Pericrocotus divaricatus Accipiter spp.
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Eurystomus orientalis Nycticorax nycticorax Haliaeetus ichthyaetus
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 0
## Cacatua goffiniana Aerodramus maximus Anthracoceros albirostris
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 0
## Picus vittatus Psilopogon haemacephalus Chalcophaps indica
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 0
## Chrysophlegma miniaceum Cuculus micropterus Nisaetus cirrhatus
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 0
## Orthotomus sericeus Pycnonotus aurigaster Mixornis gularis
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 0
## Ficedula zanthopygia Muscicapa spp. Surniculus lugubris Orthotomus spp.
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Centropus bengalensis Ploceus philippinus Prinia flaviventris Zapornia fusca
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Cecropis daurica Ploceus spp. Sternula albifrons Vidua macroura
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Aviceda leuphotes Aviceda jerdoni Aerodramus germani Psilopogon lineatus
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Cacomantis merulinus Pycnonotus plumosus Streptopelia tranquebarica
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 0
## Accipitriformes Cacomantis sepulcralis Merops spp. Acrocephalus bistrigiceps
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Lonchura spp. Centropus sinensis Gracula religiosa Pandion haliaetus
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Clanga clanga Accipiter trivirgatus Corvus spp. Gallus gallus (hybrid)
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Aethopyga siparaja Chrysococcyx xanthorhynchus Hierococcyx nisicolor
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 0
## Pycnonotus zeylanicus Treron curvirostra Lonchura leucogastroides
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 0
## Garrulax leucolophus Terpsiphone paradisi Ardea spp. Muscicapa ferruginea
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Pluvialis fulva Caprimulgus macrurus Sturnia sinensis Ducula aenea
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Myiopsitta monachus Accipiter gularis Scolopacidae Caprimulgus affinis
## 1 0 0 0 0
## 2 0 0 0 0
## 3 0 0 0 0
## 4 0 0 0 0
## 5 0 0 0 0
## 6 0 0 0 0
## Clamator coromandus Lonchura atricapilla Mycteria spp.
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 0